The BioGears code base consists of:
- Engine - A C++ based lumped parameter model
- Common Data Model - A set of generalized data classes for defining the data interfaces of a Physiology Engine Interface
- Toolkit - Various tools for execution and data manipulation, primarily written in Java
The BioGears source is structured as follows:
- bin - Contains all data and configuration files needed for execution of the BioGears Engine
- data - Contains the Microsoft Excel spreadsheets for all BioGears data sets
- lib - Contains third party libraries used by this project (see Third Party Credits for more details)
- src
- cdm - Code associated with the Common Data Model and Physiology Engine Interface
cmake - The directory where cmake will create build files
- engine - Code associated with the lumped parameter models
- controller - These classes hold data necessary by the model, control the advancement of time
- scenario - These classes help execute a BioGears specific scenario (i.e. a scenario with a BioGearsConfiguration object)
- BioGearsScenario -
- BioGearsScenarioExec -
- BioGearsScenarioInitialParameters -
- BioGears -
- BioGearsCircuits -
- BioGearsCompartments -
- BioGearsConfiguration -
- BioGearsEngine -
- BioGearsSubstances -
- BioGearsSystem -
- scenario - These classes help execute a BioGears specific scenario (i.e. a scenario with a BioGearsConfiguration object)
- systems - These classes implement the methodology for modeling and simulating
- BloodChemistry -
- Cardiovascular -
- Drugs -
- Endocrine -
- Energy -
- Environment -
- Gastrointestinal -
- Hepatic -
- Nervous -
- Renal -
- Respiratory -
- SaturationCalculator -
- Tissue -
- equipment
- AnesthesiaMachine -
- ECG -
- Inhaler -
- gui - Code associated with the Toolkit GUI
schema - The xsd data definitions used by the CDM
- sdk - Example code and scripts for the Toolkit and Software Development Kit
- utils - Various utilities used in validation
- verification - Extract the verification files into this directory. You can get more detail here.
Building
Required Tools
CMake
We generate project files with the CMake system to support a cross compliler independent code base. The BioGears team primarily develops with Visual Studio 2015, but we have created and tested the following cmake configurations:
The following projects will be generated:
- ALL_BUILD - Utility project for building
- BioGearsEngine - A shared library project of the implementation of the BioGears Physiology Engine
- BioGearsEngineJNI - A shared library exposing the BioGearsEngine through the Java Native Interface (JNI)
- BioGearsEngineTest - An optional shared library project that contains unit test classes associated with testing the BioGears methodology
- BioGearsScenarioDriver - The command line scenario executable provided in the Toolkit
- CommonDataModel - This is a shared library project that builds all code associated with the Common Data Model
- CommonDataModelJNI - A shared library exposing the CDM through the Java Native Interface (JNI)
- CommonDataModelTest - A shared library project that contains unit test classes associated with testing the CDM implementation
- DataModelBindings - This shared library project generates and builds C++ code from the CDM xsd files via Code Synthesis
- log4cpp - A shared library project of the log4cpp library
- UnitTestDriver - An executable harness for running unit tests from the two unit test projects
- ZERO_CHECK - Makes sure that project files are up to date with CMAKE files
The required projects to getting a usable C++ shared library are:
- log4cpp
- DataModelBindings
- CommonDataModel
- BioGearsEngine
Other projects are all optional and are either unit test, tools, or Java interfaces projects.
Ant
You can download and install Apache Ant from here. For this installation, choose the zipped archive and extract the files to a new directory. Ensure you add the Apache Ant bin directory to the path.
Eclipse
Eclipse is used to aid in Java code development. In the future we will support C++ project for use with Eclipse.
Java Development Kit (JDK)
We have developed a set of testing tools as well as our GUI in Java. We provide a very simple JNI interface to the C++ based engine. In the future we plan to expand the functionality provided in the Java Engine Wrapper.
Documentation Generation Tools
BioGears uses Doxygen to generate all documentation from both in-code comments as well as markdown files. We also have a set of custom Java based preprocessors to help generate various tables and other documentation into markdown files. The markdown files include both math formulas as well as bibliography citations. In order to generate the BioGears documentation you will need to have the following tools installed
- A LaTeX compiler such as
- MikTeX or proTeXt for windows
- MacTeX for MacOSX
- TeX Live for Unix/GNU/Linux
- A Perl compiler
- Ghostscript
- Doxygen
Currently there is only a windows based script for generating documentation and it is specialized for use in the BioGears code repository. For more information on using this script come to our forums.
Eclipse
The following Eclipse projects are included in the source:
- BioGearsEngine - A Java interface to the BioGearsEngine
- BioGearsEngineGUI - The Java Swing GUI provided in the Toolkit
- CommonDataModel - A Java implementation of the Common Data Model, also contains the Comparison and Graphing utility.
The BioGearsEngineGUI project require the the C++ JNI projects be built in order to execute a scenario.
Recommended execution/debugging settings from Eclipse:
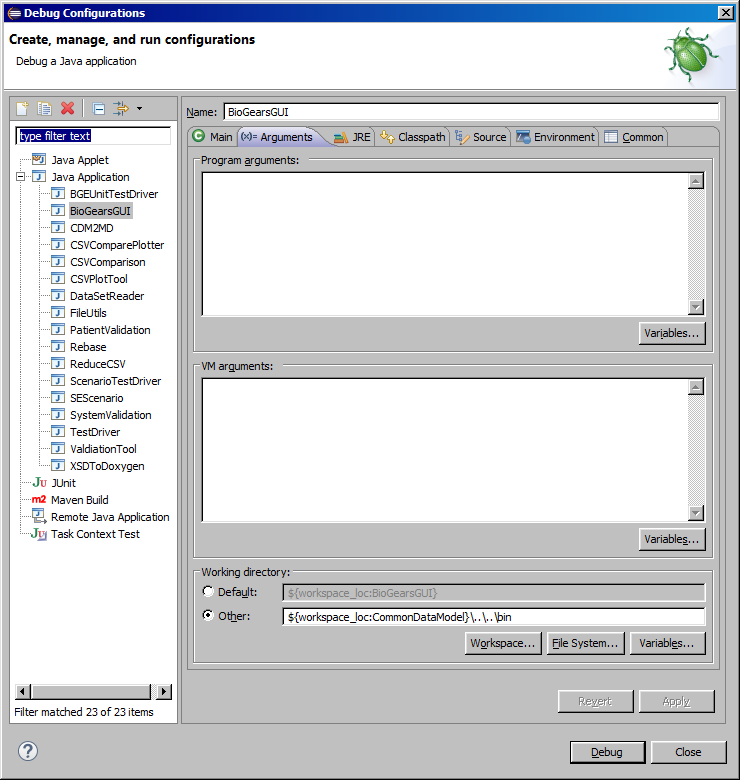
The following Java classes have main methods and can be executed:
- BioGearsEngine - Executes a scenario through a Java wrapper
- BioGearsGUI - Executes the GUI
- DataSetReader - Reads the data/BioGears.xlsx and generates all patient and substance XML files from data in the spreadsheet
- CSVPlotTool - Generates image plots for a results file. Used via the Toolkit GraphResults.bat.
- CSVComparison - Compares two results files and generates a report. Used via the Toolkit CompareResults.bat.
Building with Visual Studio
Create the Visual Studio solution and project files by running the cmake target via ant. By default, CMake will generate 32-bit Visual Studio 2015 project files in the src/cmake/msvs14x32 directory.
Note: 64-bit project files can be generated by specifying -Darchitecture=x64 in the ant command, but we are still testing this build and currently do not recommend using it.
All builds copy their compiled targets to /bin/release or /bin/debug depending on MSVC build configuration. Execution and debugging working directories should be set to the bin directory.
Recommended execution/debugging settings for Visual Studio:
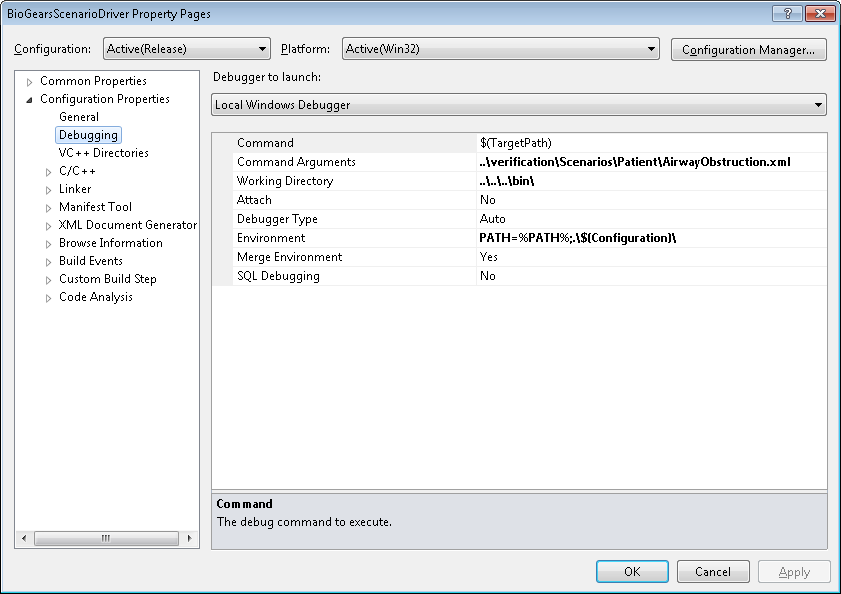
Building with GCC on Windows
BioGears can be built with GCC on Windows using MinGW-w64. Note that MinGW-w64 is NOT the same as MinGW. It is a separate version that branched off from MinGW in 2005 and contains the necessary C++11 threading libraries need to support multiple instances of a BioGears Engine within an application. Download and install MinGW-w64 using the indicated settings:

Create an environment variable MINGW_HOME which points to MinGW-w64's installation directory, e.g. C:/mingw-w64/i686-5.1.0-posix-dwarf-rt_v4-rev0/mingw32
When running ant commands, append -Denv=mingw to specify that the command should be run for the MinGW environment. For example, compile BioGears by running the ant commands ant cmake -Denv=mingw and ant compile -Denv=mingw in the src directory.
Building with Xcode on Mac OS X
Xcode
Download and install Xcode from the app store. After the installation finishes, install the Xcode command line tools by running the terminal command xcode-select --install.
Required tools
Make sure the following tools are installed:
- ant
- CMake
- Java JDK 8
An easy way to install ant and CMake is with Homebrew. First install homebrew by running the command ruby -e "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/master/install)". Next install ant and CMake by running the commands brew install ant and brew install cmake.
Build Xerces
Navigate in a terminal window to the xerces-3.1.2 directory in the BioGears lib directory. Build Xerces by running ./configure --disable-threads --disable-network --enable-transcoder-macosunicodeconverter --disable-pretty-make CXXFLAGS=-O3 CFLAGS=-O3 and then make.
After Xerces builds successfully, we have to set the dylib's install path. Navigate to the xerces-c-3.1.2/src/.libs directory, and run the command install_name_tool -id @rpath/libxerces-c-3.1.dylib libxerces-c-3.1.dylib. This sets the install path of the dylib to so that the BioGears executable will search the current directory when attempting to load it. Run the command otool -D libxerces-c-3.1.dylib to make sure the install path has been set correctly, this should output @rpath/libxerces-c-3.1.dylib.
Building and Running BioGears in Xcode
Navigate in a terminal window to the BioGears src directory and run ant cmake -Denv=xcode. This will generate debug and release Xcode project files in src/cmake/xcode.
Open the BioGears.xcodeproj file in Xcode. Select Product > Scheme > Edit Scheme and select Run on the left column. Set the following configuration options:
- Set the executable to BioGearsScenarioDriver if it isn't already.
- Set the build configuration to Debug or Release depending on the desired mode.
- On the arguments tab, add an entry to "Arguments Passed On Launch" which specifies a path to a scenario to run (e.g. verification/Scenarios/Basic/Basic1.xml).
- On the options tab, check "Use custom working directory" and set it to the BioGears bin folder.
Close the dialog and click the Run button to build and launch the BioGears scenario driver.
Building and Running BioGears from the Command Line
Navigate in a terminal window to the BioGears src directory and run ant cmake -Denv=xcode. This will generate debug and release Xcode project files in src/cmake/xcode.
To build BioGears from the command line, open the generated xcodeproj file in Xcode. Opening the project in Xcode will force the project schemes to generate, these are necessary for command line builds. After Xcode loads the project it can be closed. Run the command ant compile -Denv=xcode in BioGears's src directory to build.
After BioGears is built, navigate to the bin directory and launch the scenario driver from there, specifying the scenario you want to run as the first parameter (e.g. ./Release/BioGearsScenarioDriver verification/Scenarios/Basic/Basic1.xml).
Running the GUI from Eclipse
In addition to configuring Eclipse to run BioGears as described in the Eclipse section, we also have to specify the absolute path to the BioGearsJNI dylib so the engine can load it. Open BioGearsEngine.java and locate the runScenario method. Replace the path in the System.load(...) call with the absolute path to the BioGearsEngineJNI.dylib file (e.g. System.load("/Users/name/%BioGears/bin/Release/BioGearsEngineJNI.dylib")).
Now right click on the BioGearsGUI project and select Properties. Select Java Build Path, on the Source tab expand BioGearsGUI/java and set the Native library location to be the directory where the dylibs are located. If running in release mode, this will be the release directory in the BioGears bin directory.
Building with GCC on Linux
Required tools
Make sure the following tools are installed:
- ant
- CMake
- Java JDK 8
- GCC
Add a JAVA_HOME variable to point to the Java installation and add it to the system PATH. This can be done by adding the following lines to ~/.profile:
Replace the JAVA_HOME value with the correct installation directory.
Build Xerces
Navigate in a terminal window to the xerces-3.1.2 directory in the BioGears lib directory. Build Xerces by running ./configure --disable-threads --disable-network --enable-transcoder-gnuiconv --disable-pretty-make CXXFLAGS=-O3 CFLAGS=-O3 and then make.
Building BioGears from the Command Line
Navigate in a terminal window to the BioGears src directory and run ant cmake -Denv=unixMake and then ant compile -Denv=unixMake.
Cross-compiling with GCC for the Raspberry Pi
Note: This build has not been thoroughly tested.
This build is very similar to the GCC Linux build, the only difference being that the code is compiled with a version of GCC that produces ARM-compatible objects.
Compiler
Install gcc-arm-linux-gnueabihf and g++-arm-linux-gnueabihf (e.g. using sudo apt-get install gcc-arm-linux-gnueabihf etc.). In addition to this, make sure gcc 4.9 is installed.
Build Xerces
Build Xerces by running ./configure --disable-threads --disable-network --enable-transcoder-gnuiconv --disable-pretty-make CC=/usr/bin/arm-linux-gnueabihf-gcc CXX=/usr/bin/arm-linux-gnueabihf-g++ CXXFLAGS=-O3 CFLAGS=-O3 --host arm-linux and then make.
Building BioGears from the Command Line
Navigate in a terminal window to the BioGears src directory and run ant cmake -Denv=raspberryPi and then ant compile -Denv=raspberryPi.
Deploying on *nix Platforms
BioGears can be deployed as an executable or as a library to be used in other applications. Each of the deployment processes below assumes the code has already been built with ant compile -Denv=your_environment.
Mac Executable
Deploy the Mac executable by running deploy-unix-executable.sh in the BioGears src directory. This will deploy the necessary files to the BioGears deploy/toolkit directory. To launch the GUI from that directory, run BioGearsGUI.sh.
Mac SDK
Deploy the Mac SDK by running deploy-osx-library.sh in the BioGears src directory. This will deploy the necessay header files and .dylib files to the BioGears library directory. The HowTo cpp files provide examples of how to use the BioGears API from your own software. The build-osx.sh script will build all of the HowTo files and place the resulting executable in the library/bin directory.
Linux Executable
Deploy the Linux executable by running deploy-unix-executable.sh in the BioGears src directory. This will deploy the necessary files to the BioGears deploy/toolkit directory. To launch the GUI from that directory, run BioGearsGUI.sh.
Linux SDK
Deploy the Linux SDK by running deploy-linux-library.sh in the BioGears src directory. This will deploy the necessay header files and .so files to the BioGears library directory. The HowTo cpp files provide examples of how to use the BioGears API from your own software. The build-linux.sh script will build all of the HowTo files and place the resulting executable in the library/bin directory.





