Overview
Abstract
The BioGears Gastrointestinal System models the ingestion of macro-nutrients and their subsequent digestion and transportation into the Cardiovascular system. Digestion rates for each macro-nutrient are calculated by their interaction with the corresponding stomach enzyme. Although many enzymes participate in this digestion process, BioGears assumes lipase, amylase, and trypsin are the enzymes facilitating digestion of fat, carbohydrates, and proteins. Each macro-nutrient, after reacting with its digestive enzyme, is modeled in BioGears as only one substance. Fat is modeled as triacylglycerol (tripalmitin), carbohydrate is modeled as glucose, and protein is modeled by a lumped amino acids substance that represents alanine. We currently do no support the various other products produced during the digestion and absorption cycle within the GI system. Once digestion takes place, the product is moved into the small intestine chyme compartment, where it is absorbed into the vasculature through the intestinal walls via the major sodium co-transporters.
Introduction
Gastrointestinal Physiology
The Gastrointestinal tract is responsible for consuming and digesting food, absorbing nutrients, and expelling waste. The tract consists of the stomach and intestines breaking down food into usable nutrients for the body. After being eaten, food is stored in the stomach, gradually digested, then released into the gut chyme. At this point, nutrients are either quickly absorbed into the blood through the intestinal wall or further broken down prior to absorption. An overview of the digestive tract is shown in Figure 1.

System Design
Background and Scope
The Gastorintestinal System was created so that end users could supply and replenish water and substances in the body to counterbalance losses associated with exercise and clearance. It provides a low-fidelity enzyme kinetic model coupled with gastrointestinal secretions to supply the major sodium co-transporter SGLUT1. Sodium absorption is directly coupled to movement of amino acids and glucose into the vasculature. Fat is handled separately with direct absorption of triacylglycerol into the blood stream, with future work targeting an expanded lymph system to facilitate more accurate fat absorption modeling and transport. Other major ions and water have constant digestion and absorption rates. Future work to properly model the major ion transport proteins will help enhance this current implementation.
Data Flow
Reset
The stomach is initialized with configurable amounts of each macronutrient, sodium, calcium, and water.
Preprocess
Gastric Secretion
A static gastric secretion flow rate of 2.46 mL/min is used to manually move water volume from the gut tissue into the stomach.
Digestion
Digestion of nutrients in BioGears is modeled as an enzymatic process that aims to replicate product formation in the human body as nutrients travel through the gastrointestinal system. The enzyme-substrate interaction and product formation is modeled using Michaelis-Menten kinetics describing the following process:
\[\left[ E \right] + \left[ S \right] \mathbin{\lower.3ex\hbox{$\buildrel\textstyle\rightarrow\over {\smash{\leftarrow}}$}} \left[ {ES} \right] \to \left[ E \right] + \left[ P \right].\]
We model this rate formation of product as a non-linear function of substrate concentration:
\[\frac{{dp}}{{dt}} = \frac{{{V_{\max }}\left[ S \right]}}{{{K_m} + \left[ S \right]}}.\]
Then multiply by our timestep to compute total mass of product moved into the intestinal chyme:
\[\Delta P = \left( {\frac{{{V_{\max }}\left[ S \right]}}{{{K_m} + \left[ S \right]}}} \right)\Delta t\]
Where *ΔP* is in units of mass and is computed each time step given the current substrate concentration on the stomach. Rate constants were taken from experimental papers, often times from in vitro studies, and can be seen in table 1. Even though these may not be perfect numbers, the mouth to cecum transit times line up well with experimental data.
| Nutrient | Enzyme | Product | Vmax (mg/min) | Km (mg/mL) |
|---|---|---|---|---|
| Fat | Lipase | Triacylglycerol | 3.40 | 0.34 |
| Carbohydrate | Amylase | Glucose | 7.11 | 3.076 |
| Protein | Trypsin | Amino Acids | 7.1 | 0.71 |
Chyme Secretion
The human body uses gastric sections to produce bile, which has the enzymes and pH level necessary to digest the nutrients eaten into absorbable substances that fuel metabolism. This process of secretions is generally performed by the pancreas and is a function of composition and quantity of the food eaten. BioGears, during digestion, assumes full availability of these digestive enzymes. Substances are then secreted into the small intestine. Bile salts are critical in digestion and absorption of nutrients into the bloodstream and are needed to facilitate our model of absorption so BioGears mimics this functionality seen in the body.
We model this functionality similarly to an enzyme kinetics model by using a non-linear function of stomach mass to compute sodium secreted. BioGears models sodium mass transfer as a Hill-type non-linear function of total digested product in the small intestine chyme:
\[\Delta N{a_{\sec }} = \left( {\frac{{{V_{\max }}{N_{mass}}}}{{{K_m} + {N_{mass}}}}} \right)\Delta t\]
Where Nmass is the maximum total mass of a nutrient currently in the stomach. Sodium is currently the only supported ion to be included in this process, but the model could be extended to include hydrogen and bicarbonate in order to validate pH levels of the chyme.
Absorption
Absorption of digested nutrients is facilitated through sodium uptake. In a similar manner to secretion absorption is modeled as a non-linear function of product mass in the chyme, as in equation 4. Once absorbed sodium mass is computed, co-transport of glucose and amino acids begins. We assume that 1 mg of sodium can transport 2 mg of glucose or 1 mg of amino acids in our simplified model. This allows for total transit times that are in line with validated data [174]
A static absorption flow rate of 3.3 mL/min is used to manually move water volume from the gut chyme into small intestine vasculature.
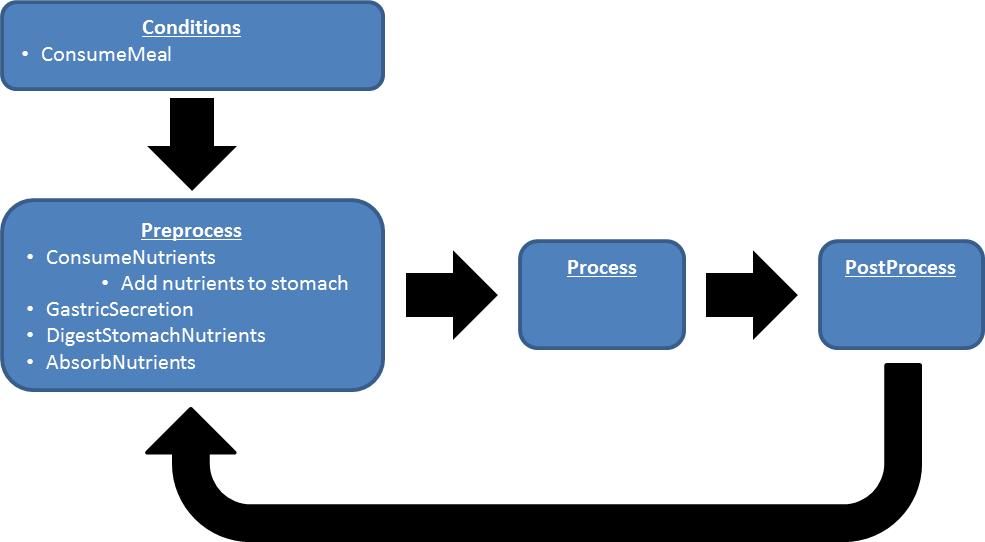
Conditions
ConsumeMeal
The ConsumeMeal condition provides a nutrition object along with an elapsed time since those nutrients were in the stomach. The GI System will then zero out the contents of the stomach and gut chyme and add the nutrient data provided in the condition to the stomach. The digestion algorithm is executed for the provided elapsed time, removing nutrient mass from the stomach and adding it to substance quantities in the gut chyme. Next, the absorption algorithm will be executed for the provided elapsed time, removing substance mass from the gut chyme and distributing that mass throughout the blood and tissues.
Actions
ConsumeNutrients
Each nutrient is optional in the ConsumeNutrients action. If a nutrient mass is provided with no accompanying digestion rate, the engine will default to the rate specified in the BioGears configuration. If a digestion rate is provided for a macronutrient, it will be combined with the current stomach digestion rate for that macronutrient by volume weighting the two rates together.
Process
There is no system specific function for Process in the Gastrointestinal System.
Post Process
There is no system specific function for Post Process in the Gastrointestinal System.
Assessments
There are no system specific assessments in the Gastrointestinal System.
Features, Capabilities, and Dependencies
Circuit

The constant secretion of fluid into the digestive tract allows for macronutrients to be slowly digested and eventually absorbed through the intestinal wall [138]. In BioGears, these macronutrients are placed into the CV system via an absorption rate that mirrors that of the human GI tract. The calculated flow rate transports substance from the small intestine C1 node to the CV system's small intestine 1 node GI-circuit. Similar transport connects the tissue and the small intestine and determines water balance and secretion rate. The digestion rate of macronutrients is heavily dependent on the nature of the source food, as the digestive tract needs to free substances from the rest of the food's molecular structure before it can be absorbed [398]. Fats spend several hours being emulsified by the small intestine and are then quickly absorbed once exposed to pancreatic enzymes [138]. Many of the small ions are quickly absorbed with active transport in a manner similar to the Renal system [138]. An exception to this is calcium, which is carefully regulated by parathyroid hormone to ensure that the body's requirements are met [138]. Water is absorbed via the osmotic gradient that either naturally exists between the gut chyme and the blood or that is created by the active transport of substances across the intestinal wall [138].
Dependencies
The Gastrointestinal System is not dependent on any other system, but other systems are dependent on GI to provided and replenish substances that they use.
Assumptions and Limitations
Currently there is no modeling of defecation or the lower portion of the digestive tract. This means that all food that is ingested will eventually be absorbed. The current digestion methodology is rudimentary and assumes all digestive rates are constant and move with the water flow after leaving the stomach. Active transport is used across cell walls with no diffusion currently implemented. Contraction of the small intestines to allow for transport and defecation is also not modeled.
Results and Conclusions
Validation - Resting Physiologic State
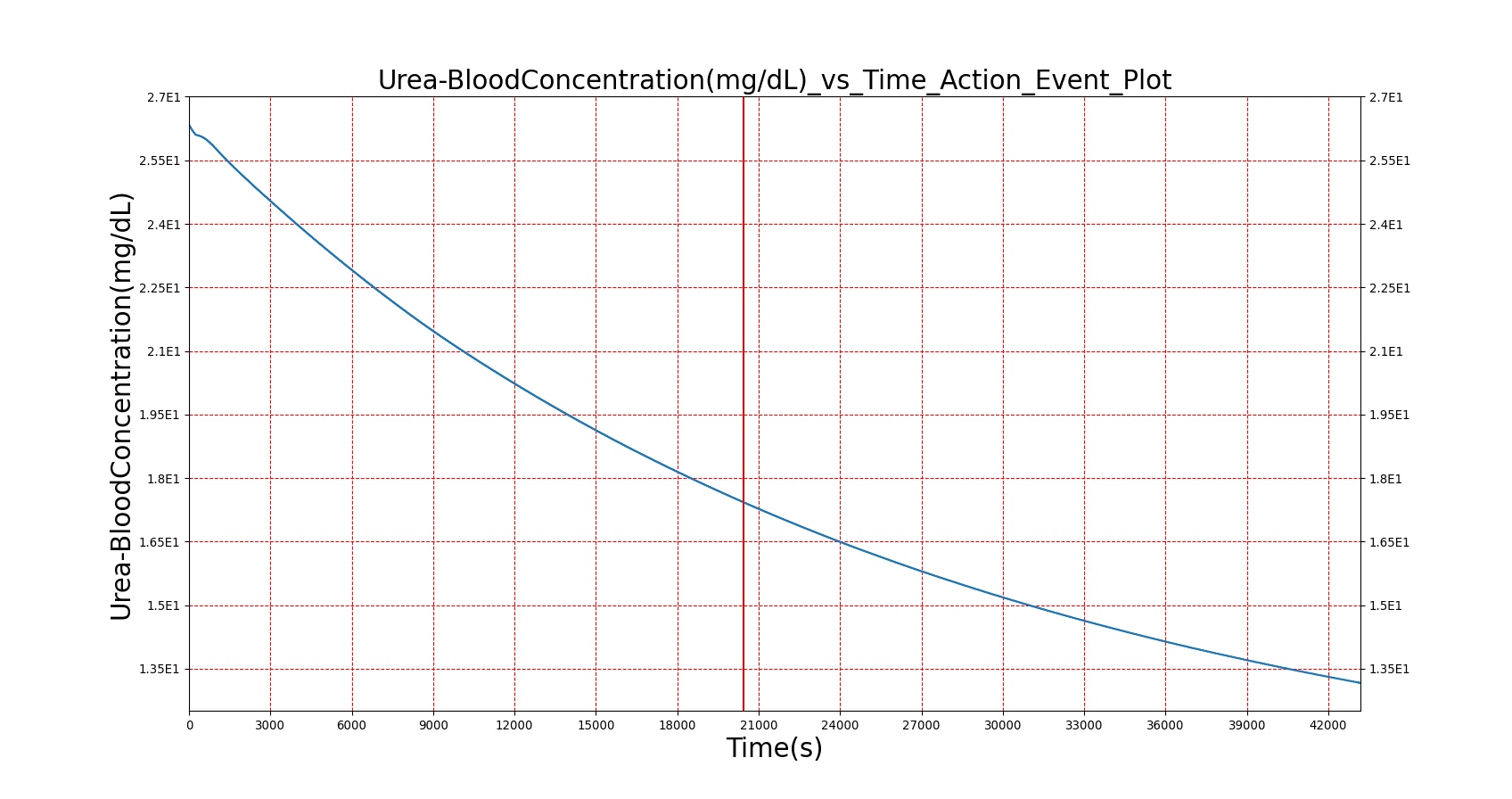
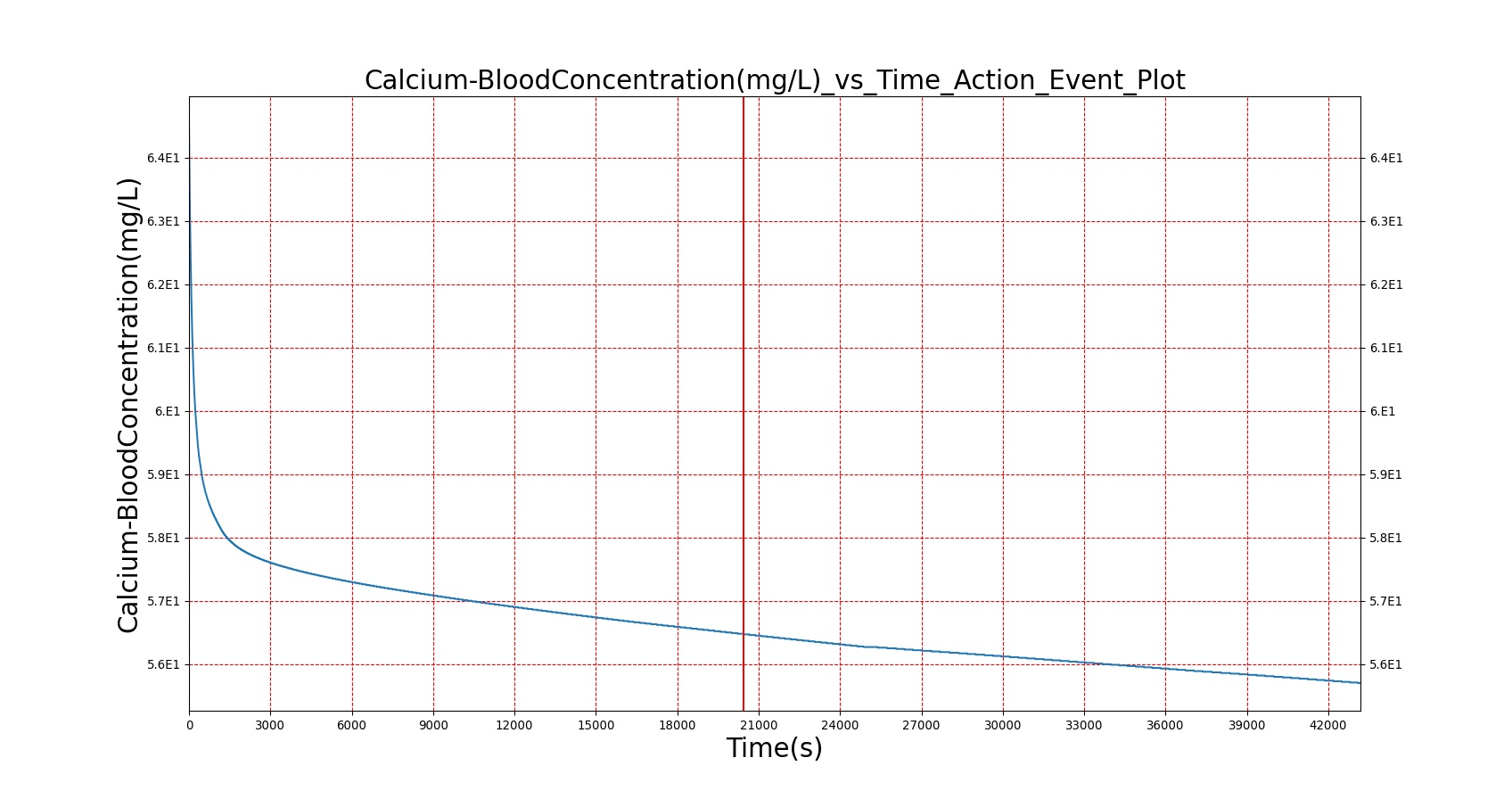
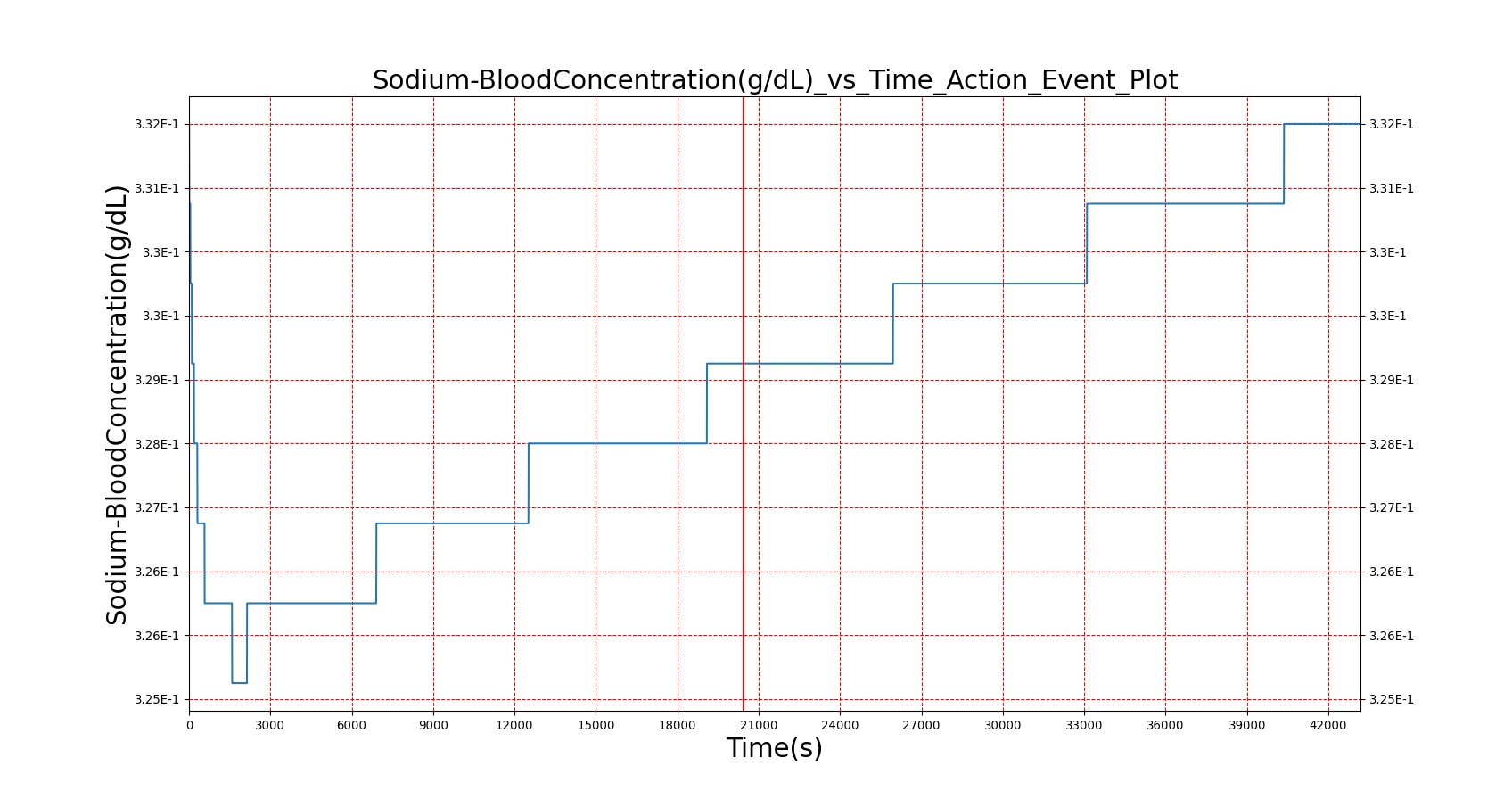
The BioGears Gastrointestinal System was validated qualitatively by observing the relative changes in the substance and nutrient masses between the stomach contents and the gut contents. The transport to the CV system was then shown by demonstrating an increase in substance concentration in the blood. Concentrations are not solely a function of GI; other systems are potentially using substances in the blood.
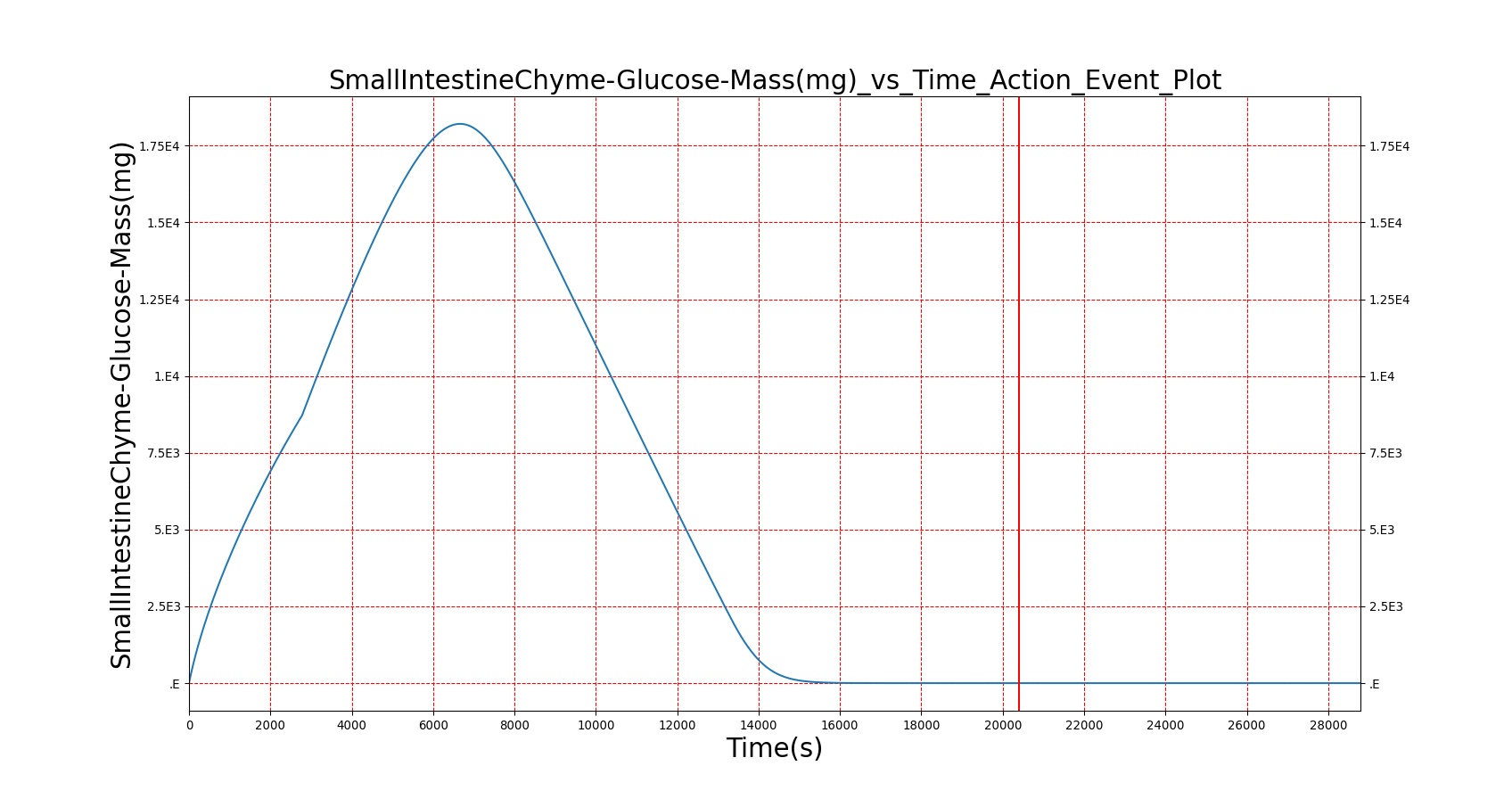
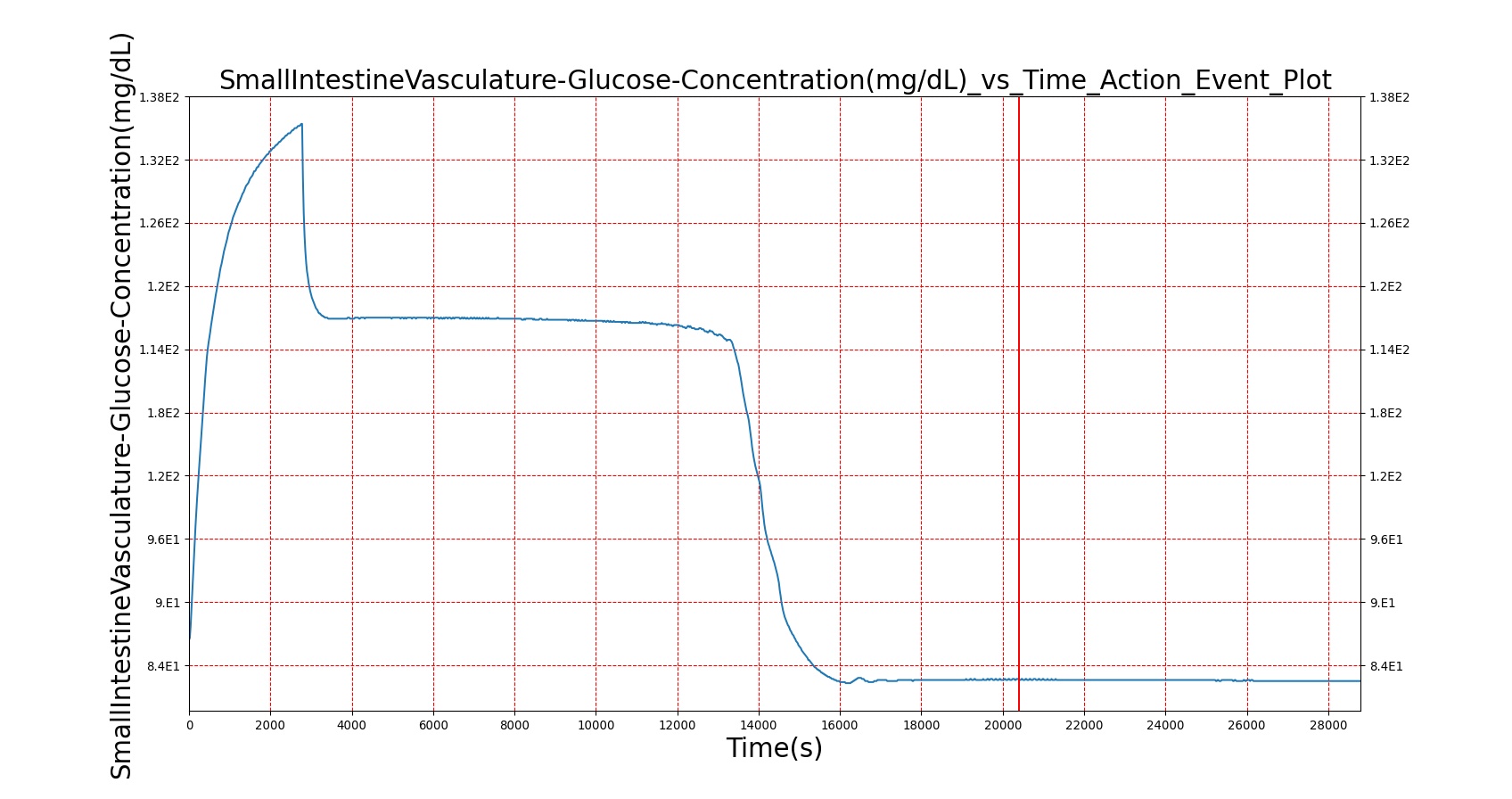
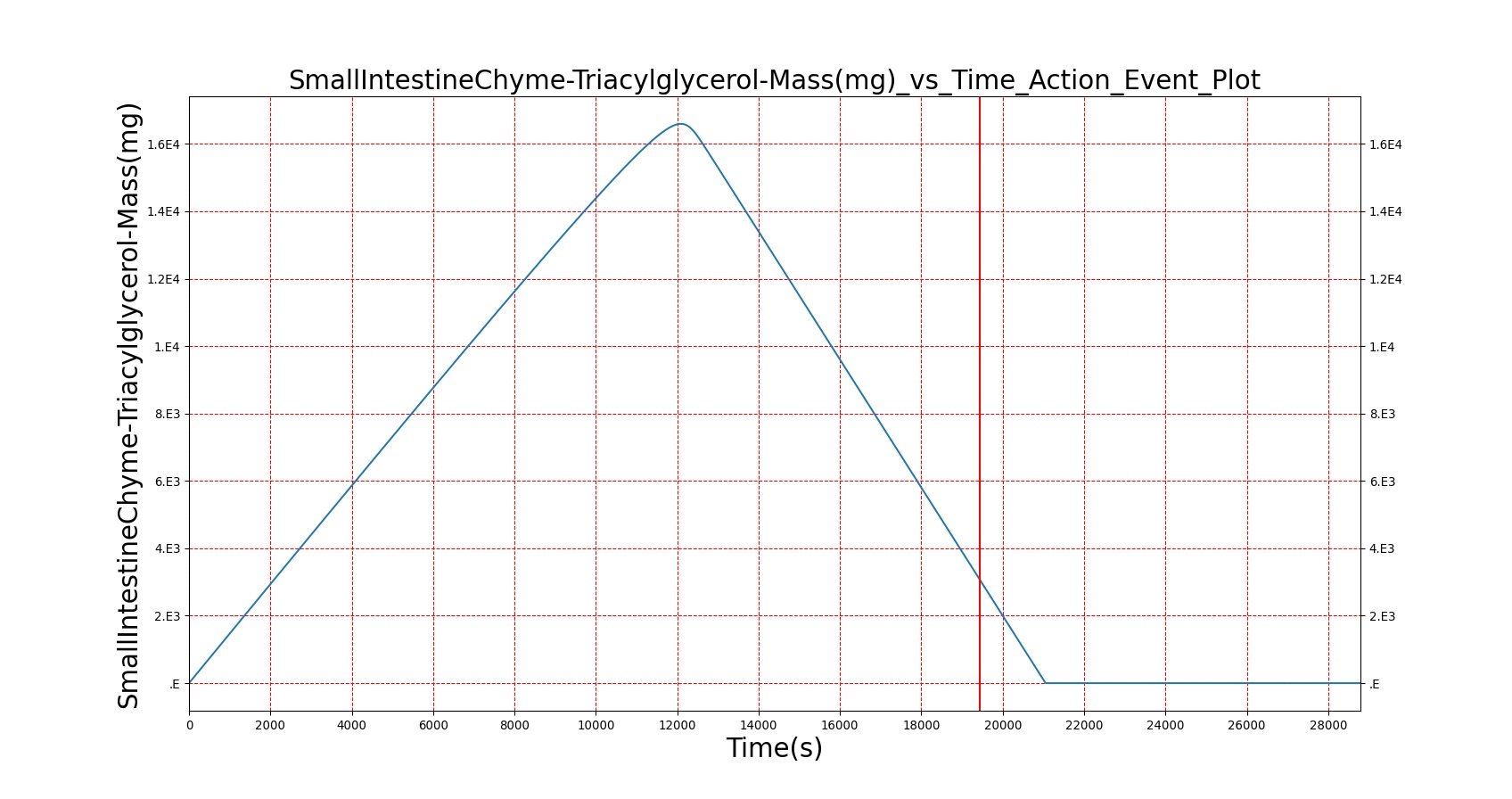
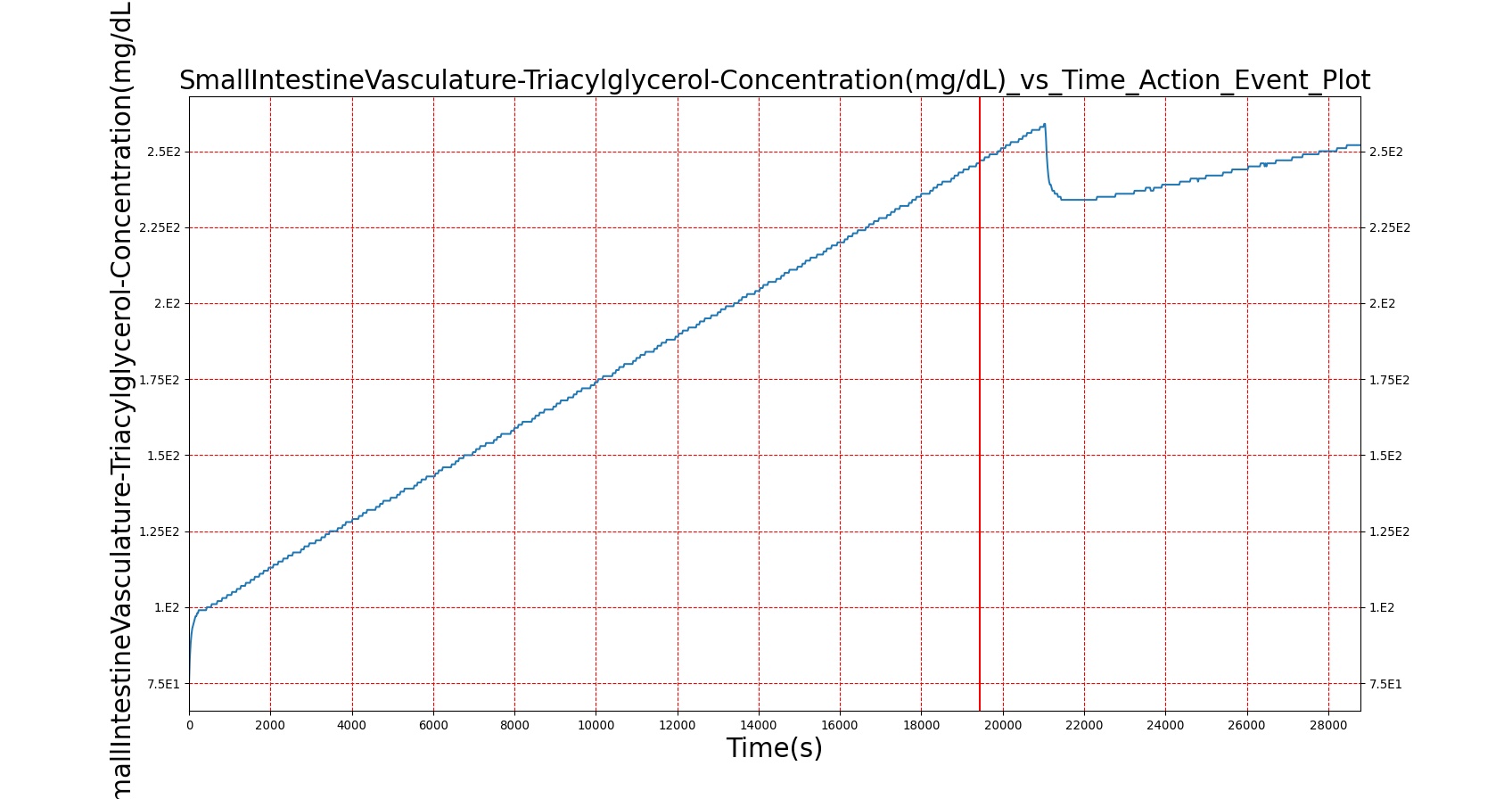
| Macronutrient/Substance | Stomach Mass(g) vs. Time(s) | Gut Mass(g) vs. Time(s) | Blood Concentration (ug/mL) vs. Time (s) |
|---|---|---|---|
| Carbohydrate/Glucose | 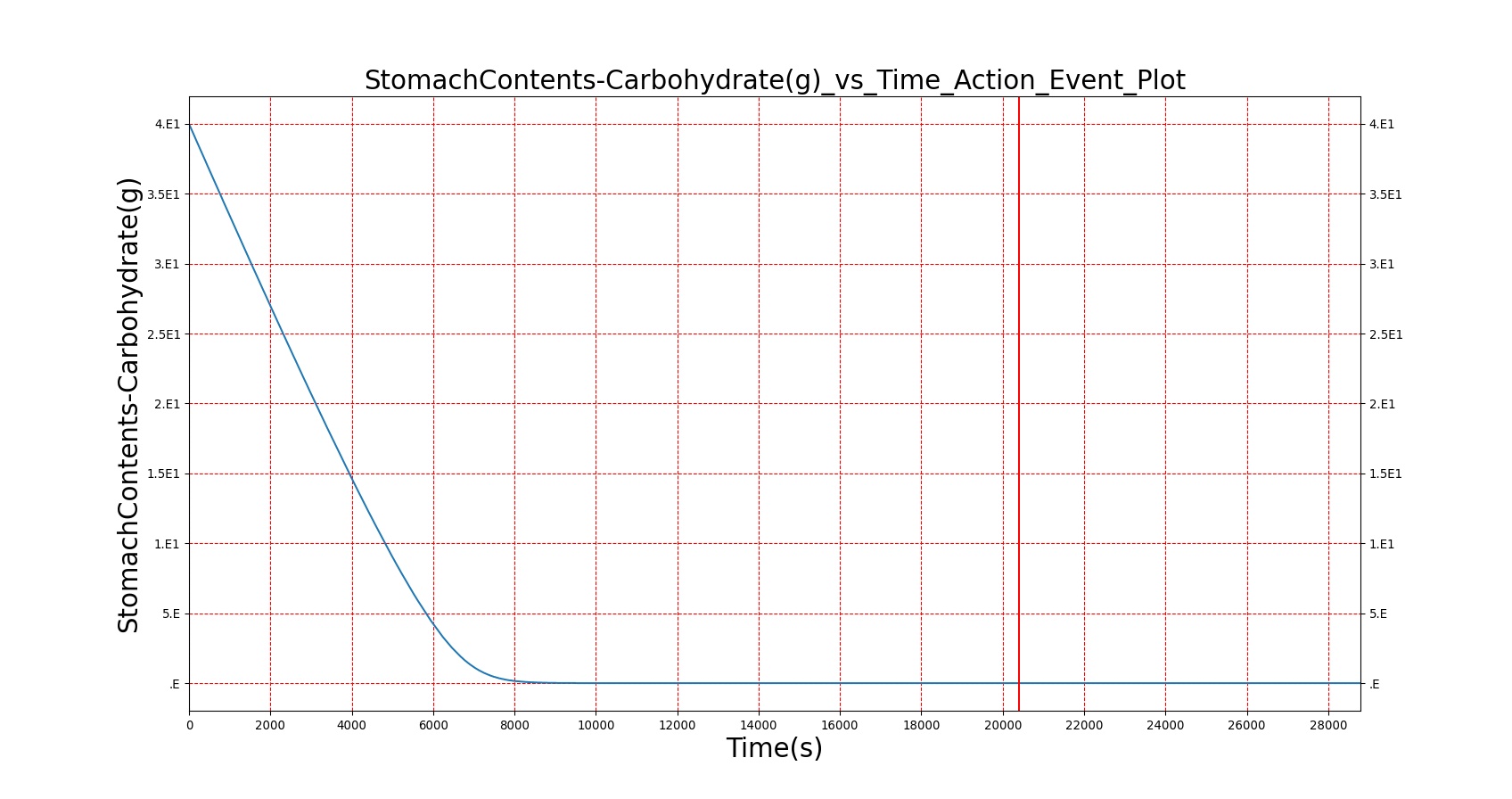
| 
| 
|
| Fat/Triacylglycerol | 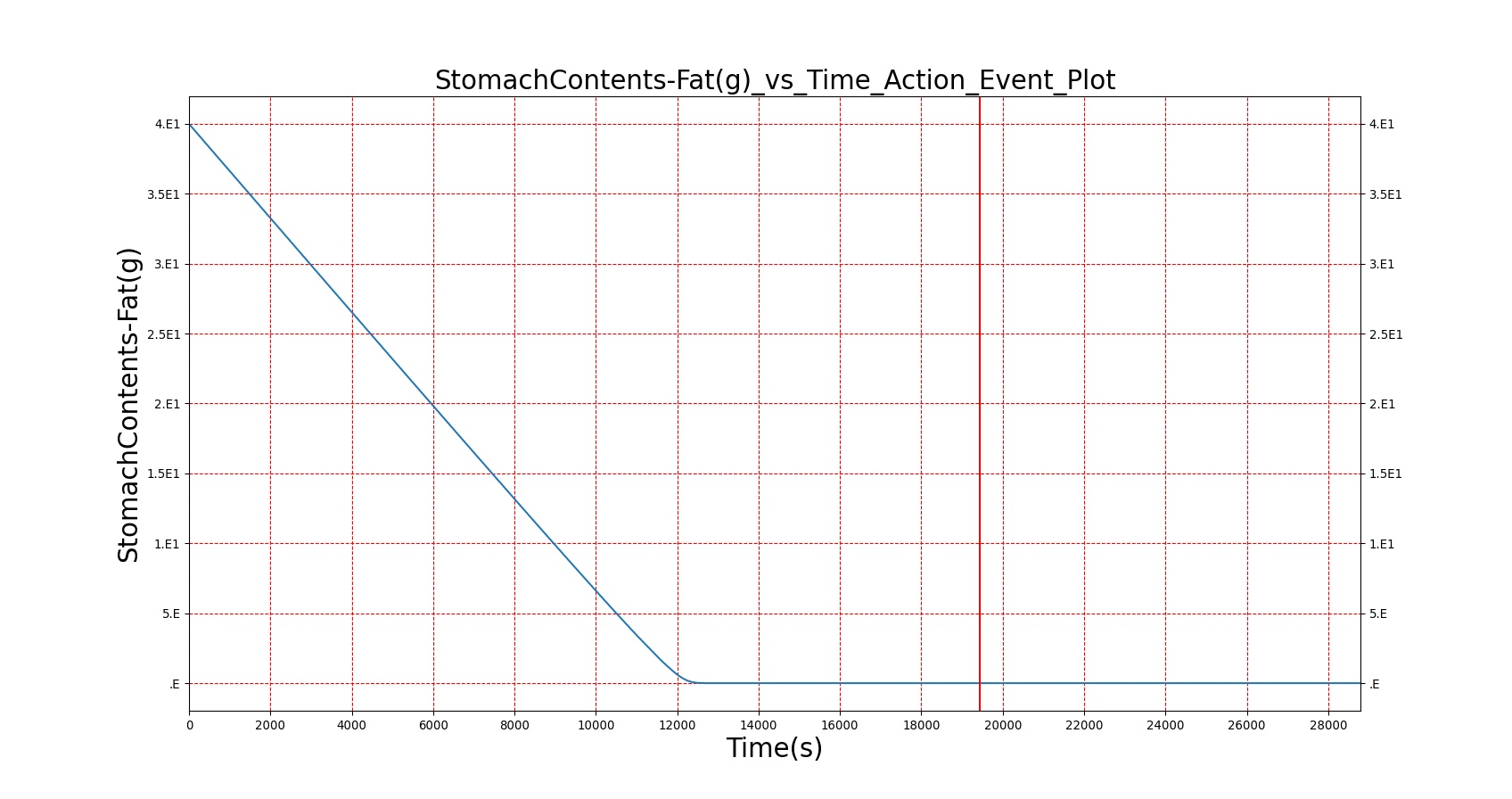
| 
| 
|
| Protein/AminoAcids | 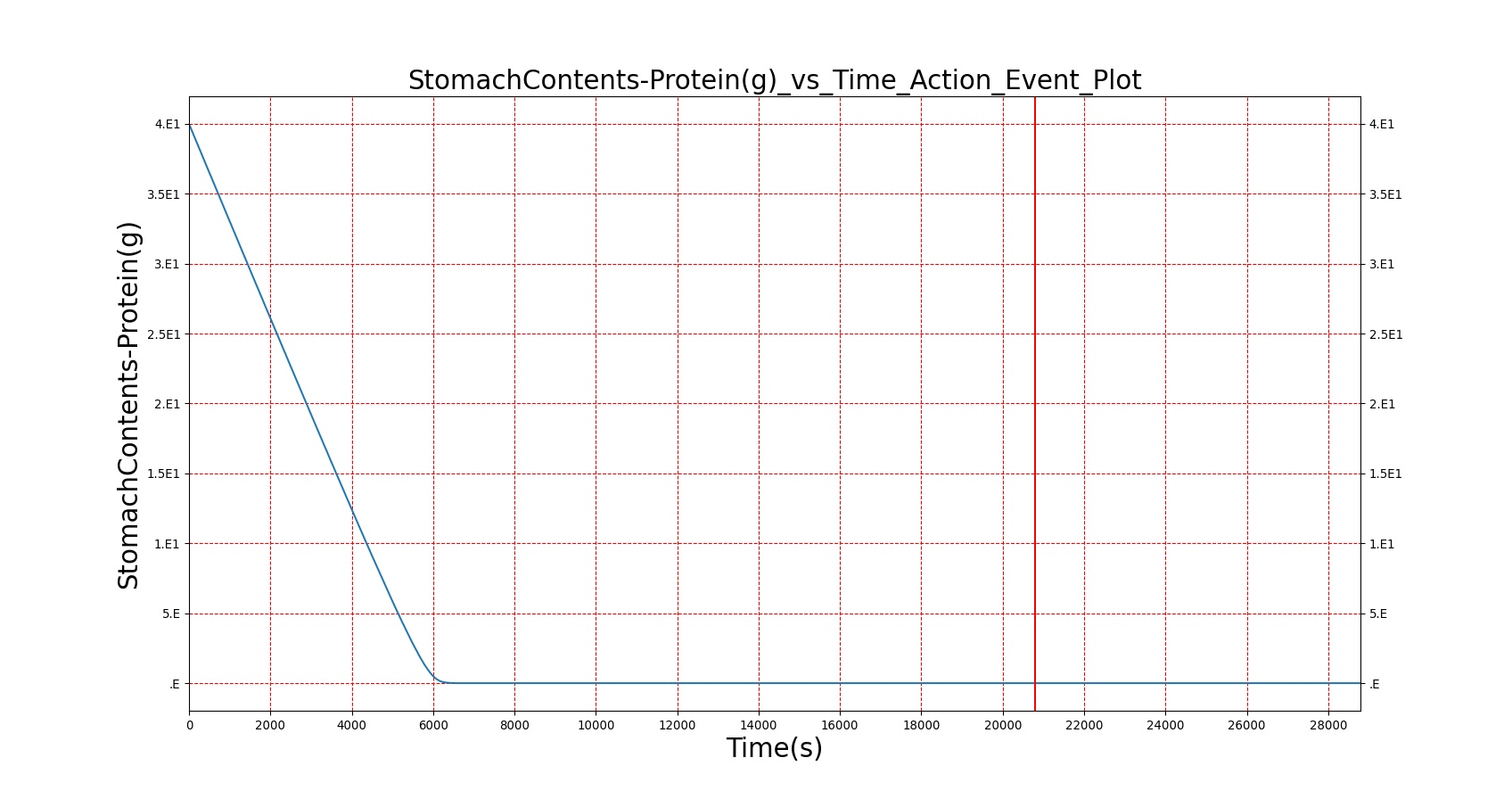
| 
| 
|
| Calcium | 
| 
| 
|
| Sodium | 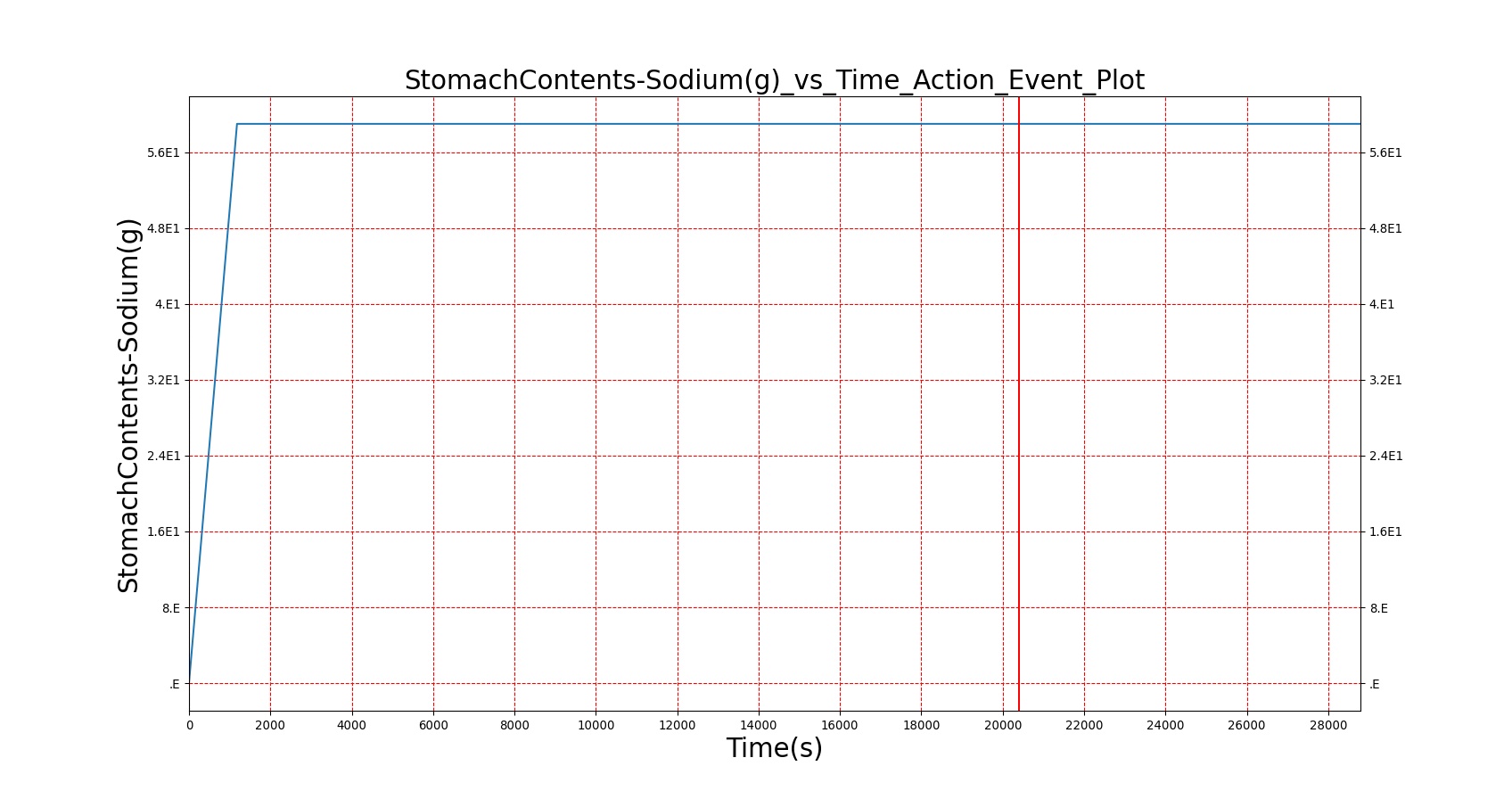
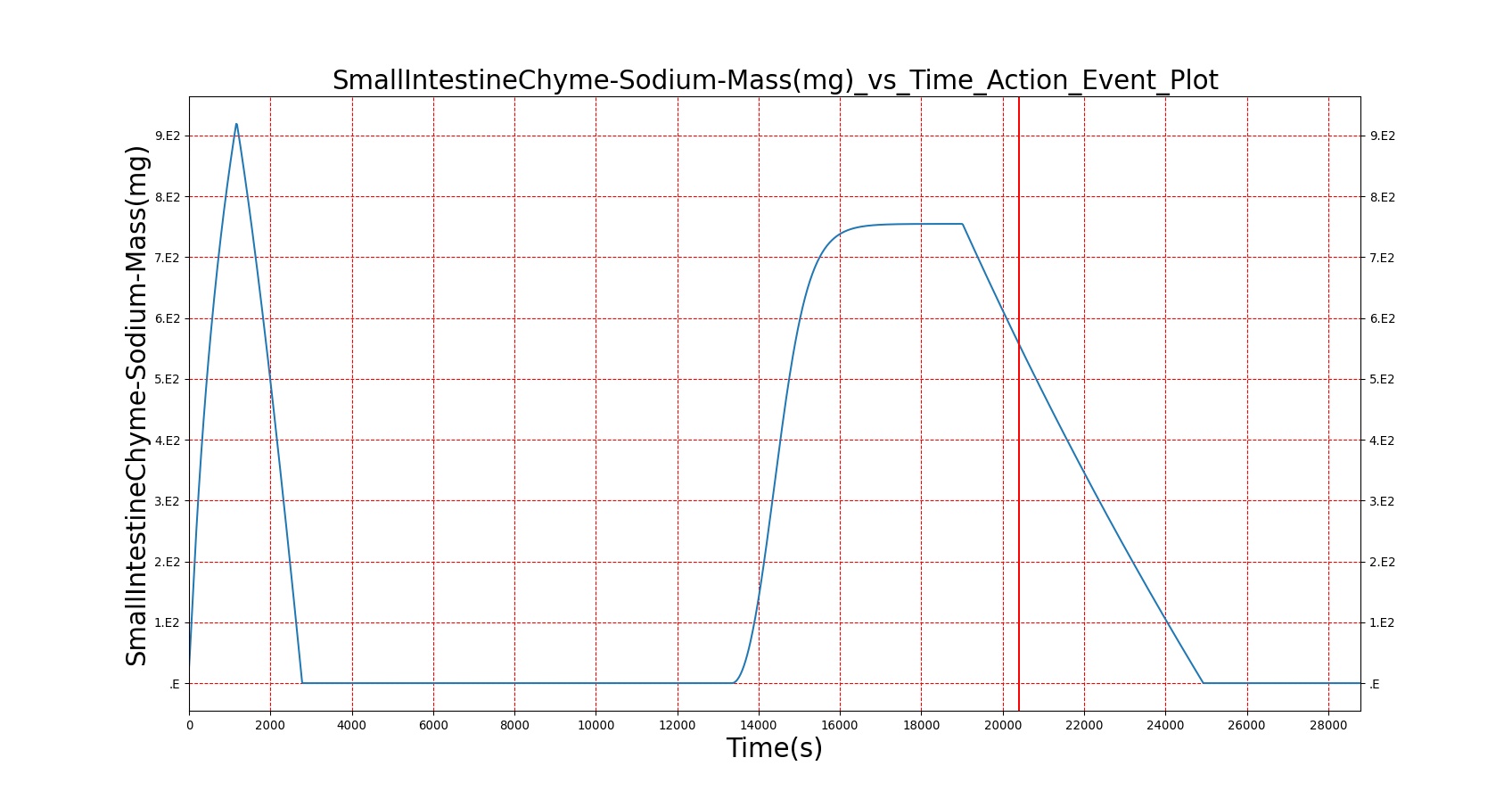
| 
| 
|
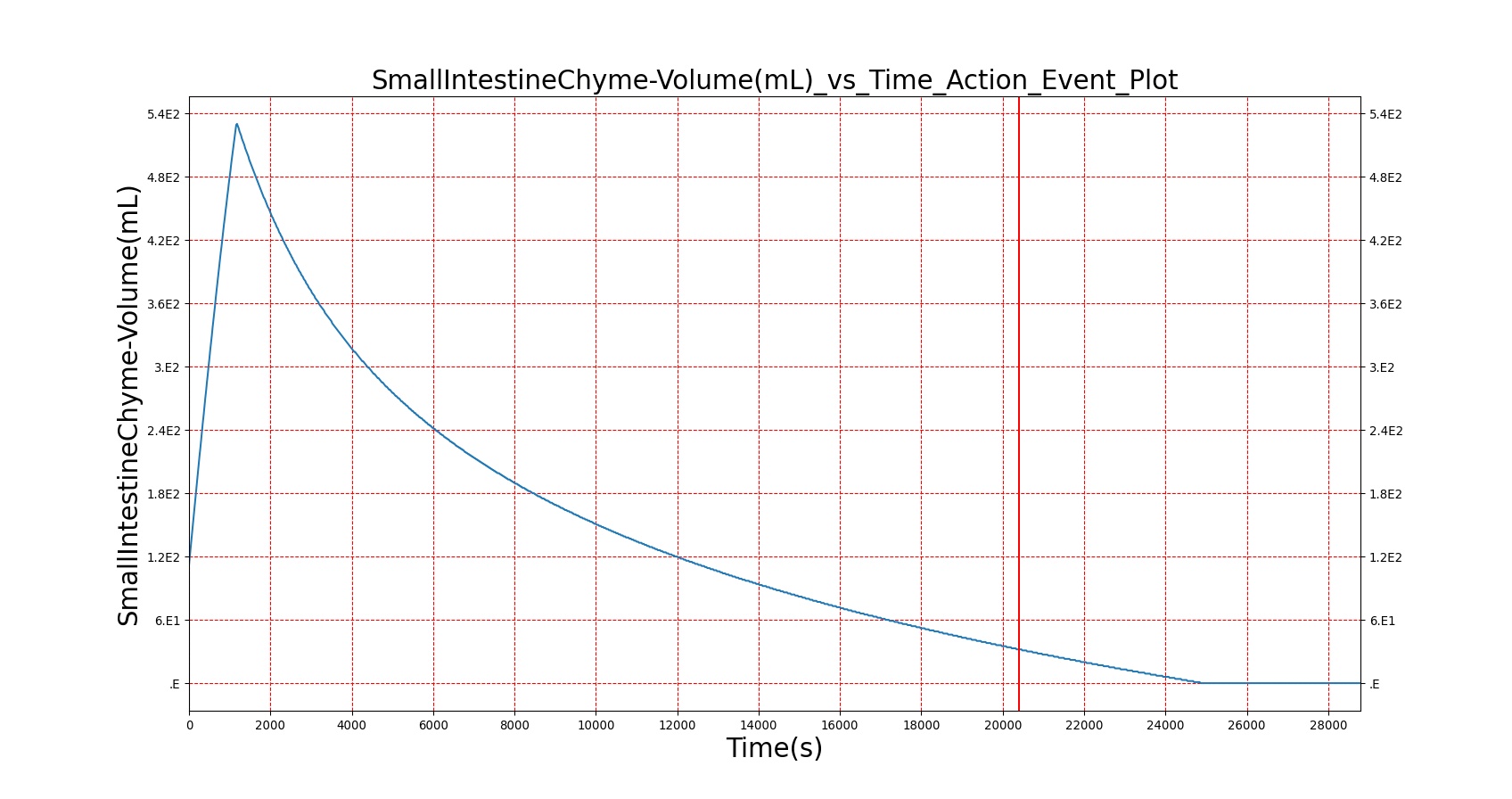
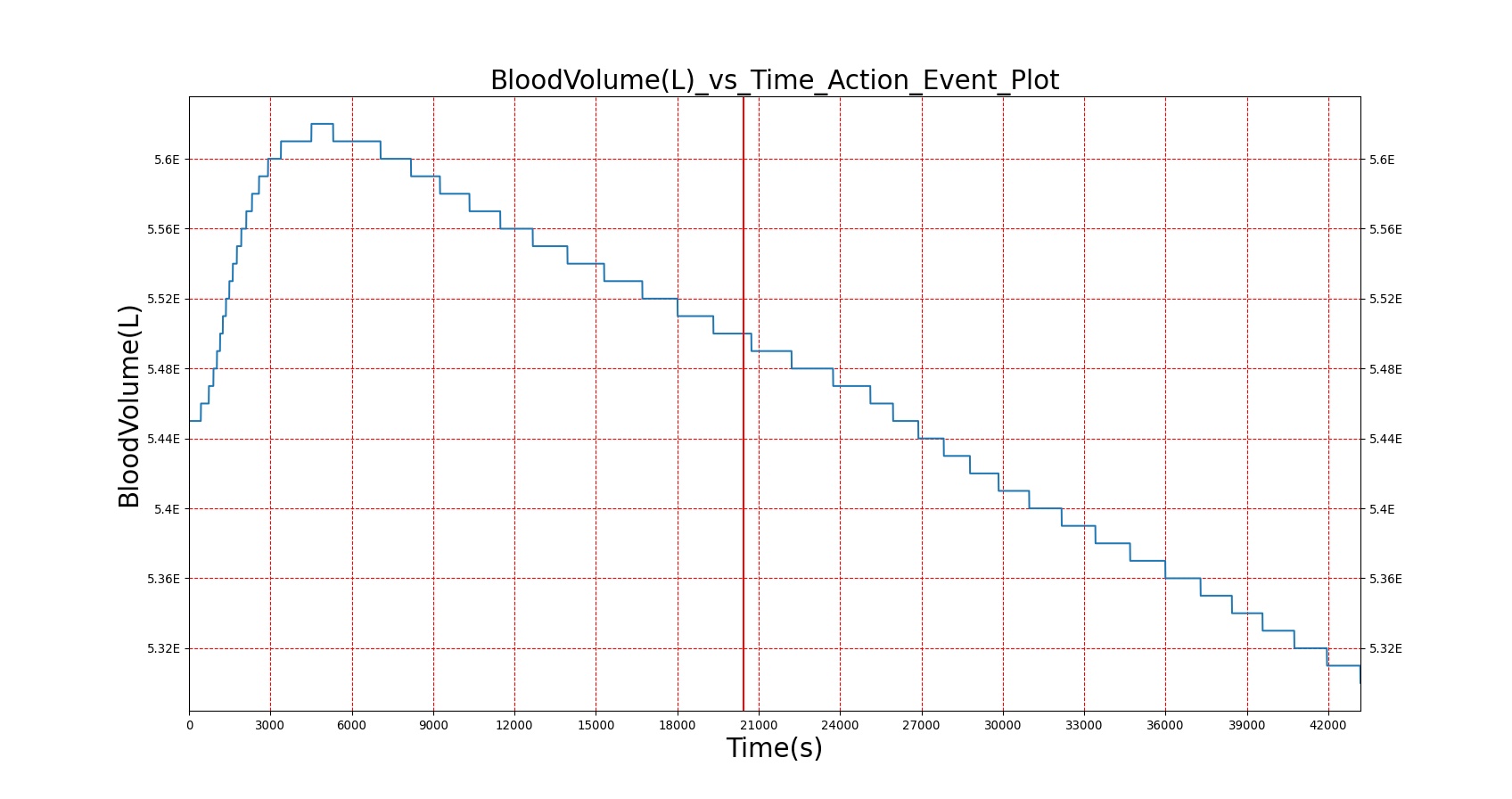
| Water | 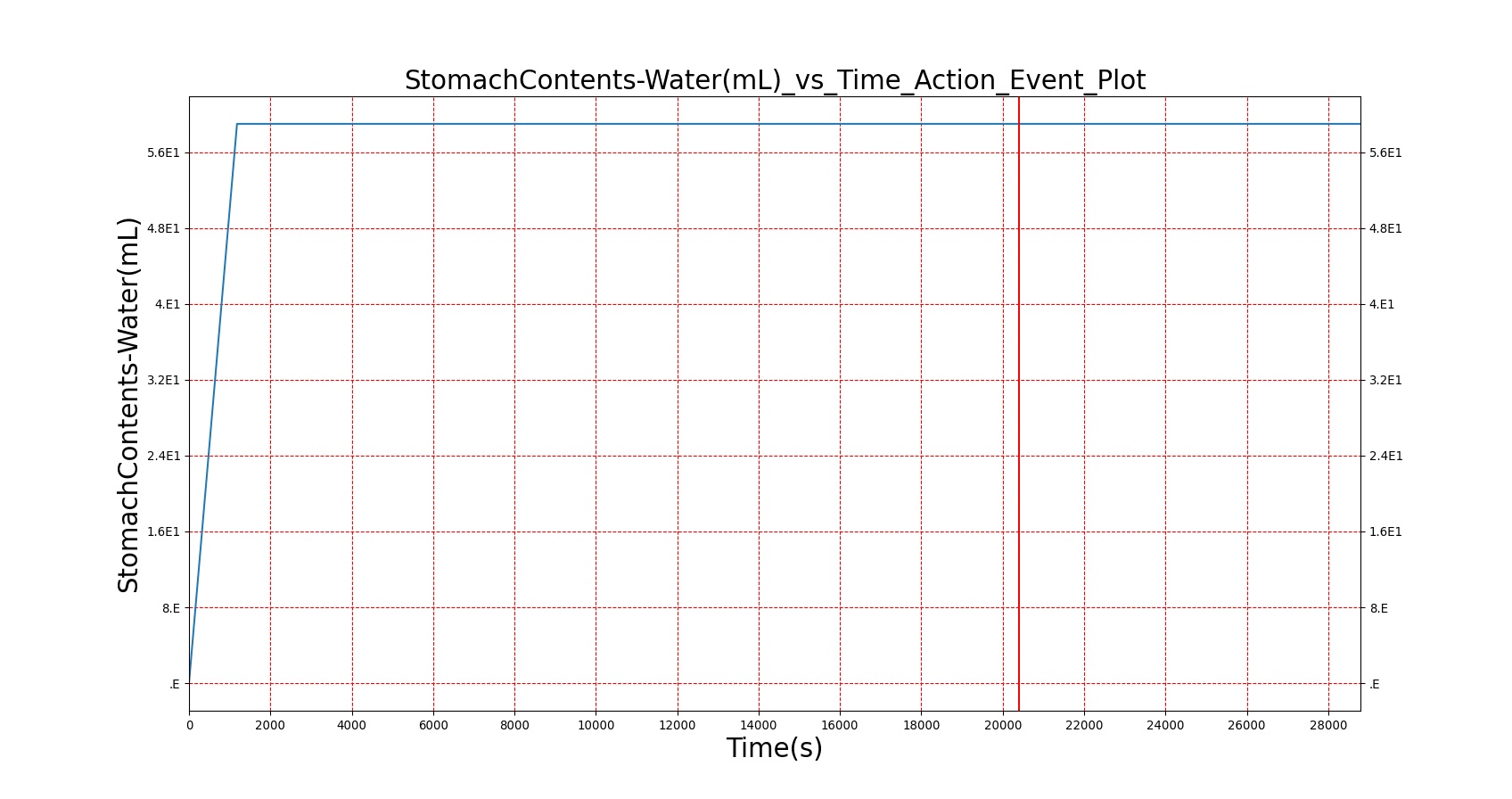
| 
| 
|
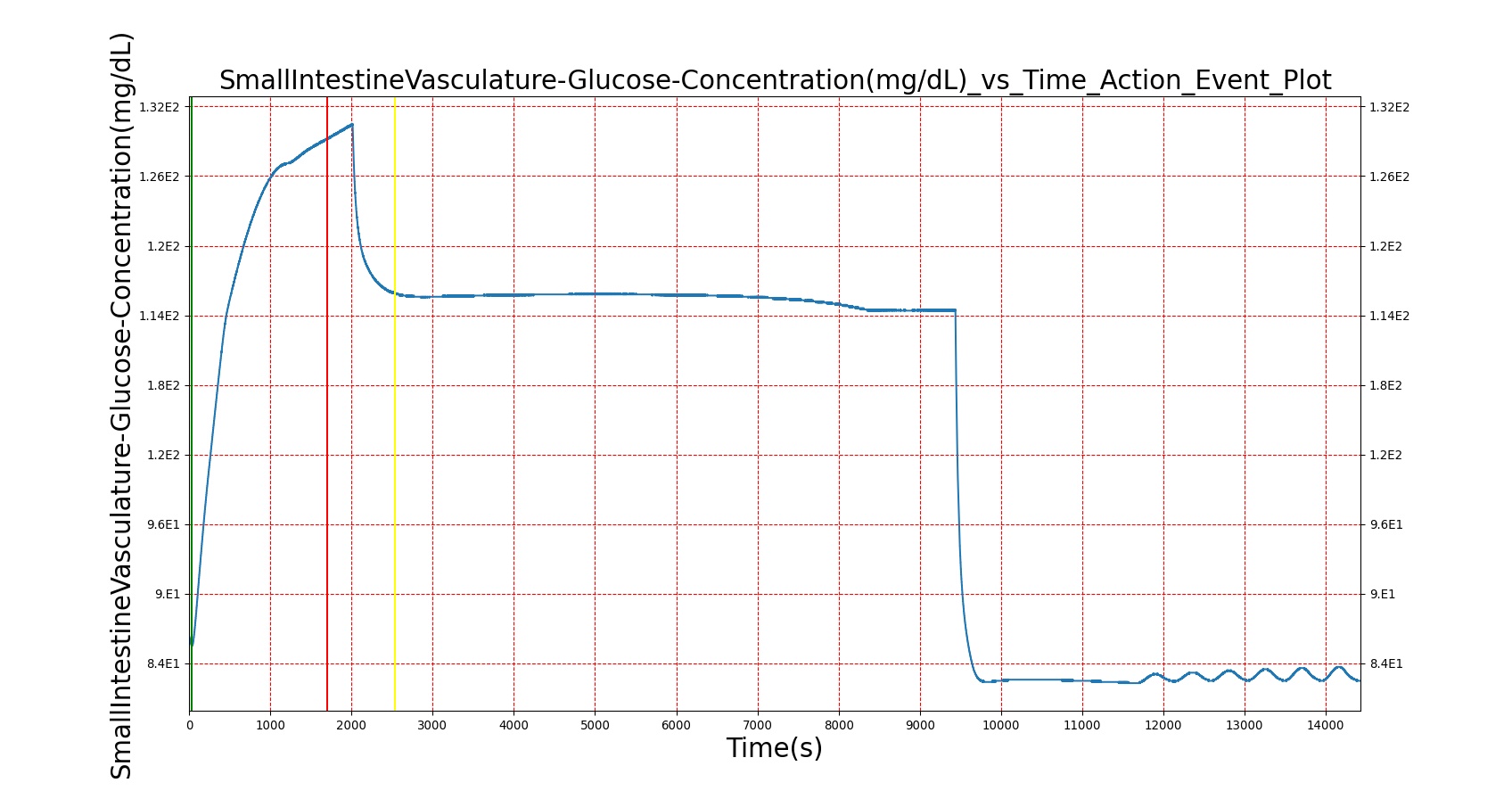
As seen in Table 2, the macronutrient masses in the stomach are depleted based on different digestion rates and eventually reach zero. The center column of plots shows the associated mass of substances in the gut chyme increase as the nutrients are removed from the stomach. Concentrations are provided in the far right column demonstrating the effect of the increasing mass on the blood concentrations. These overall trends meet the expectations of the model performance.
Validation - Actions
Consume Meal Standard Validation
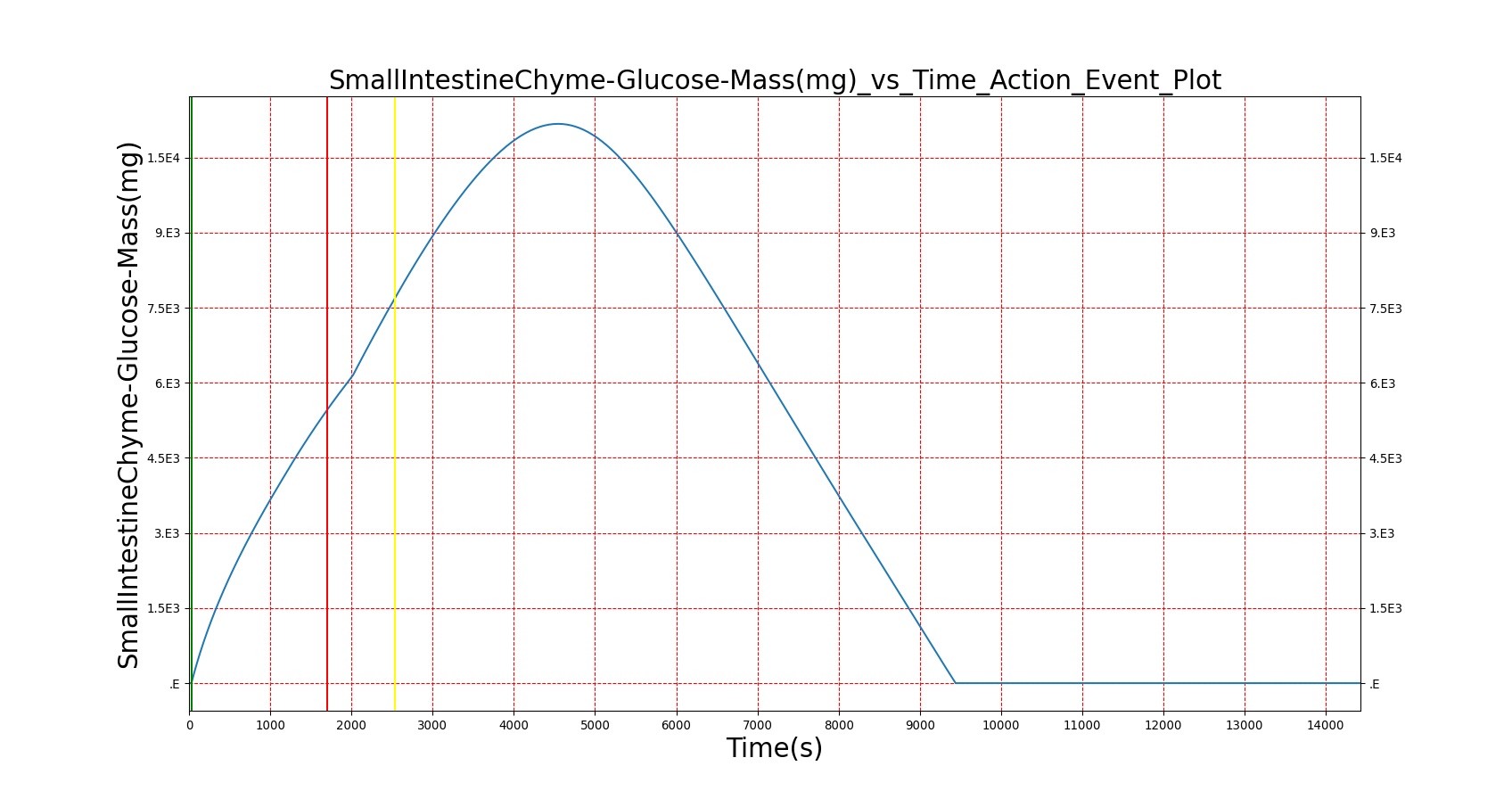
Mouth to cecum transit times are validated against experimental data [174]. Total transit is on the slow end of reported ranges of patients ingesting a standard meal but still within the deviation found in the study.
| Actions | Notes | Occurrence Time (s) | Sampled Scenario Time (hr) | Gastric emptying time (carbs) (min) | Mouth-to-Cecum Transit Time (min) | Blood Glucose Concentration return time (min) |
|---|---|---|---|---|---|---|
| — | Give a meal to the patient: 26g carbohydrate, 6g protein, 30g fat | 30s | 4hr | 40-90 min [174] | 64-144 min [174] | 120-180 min [314] |
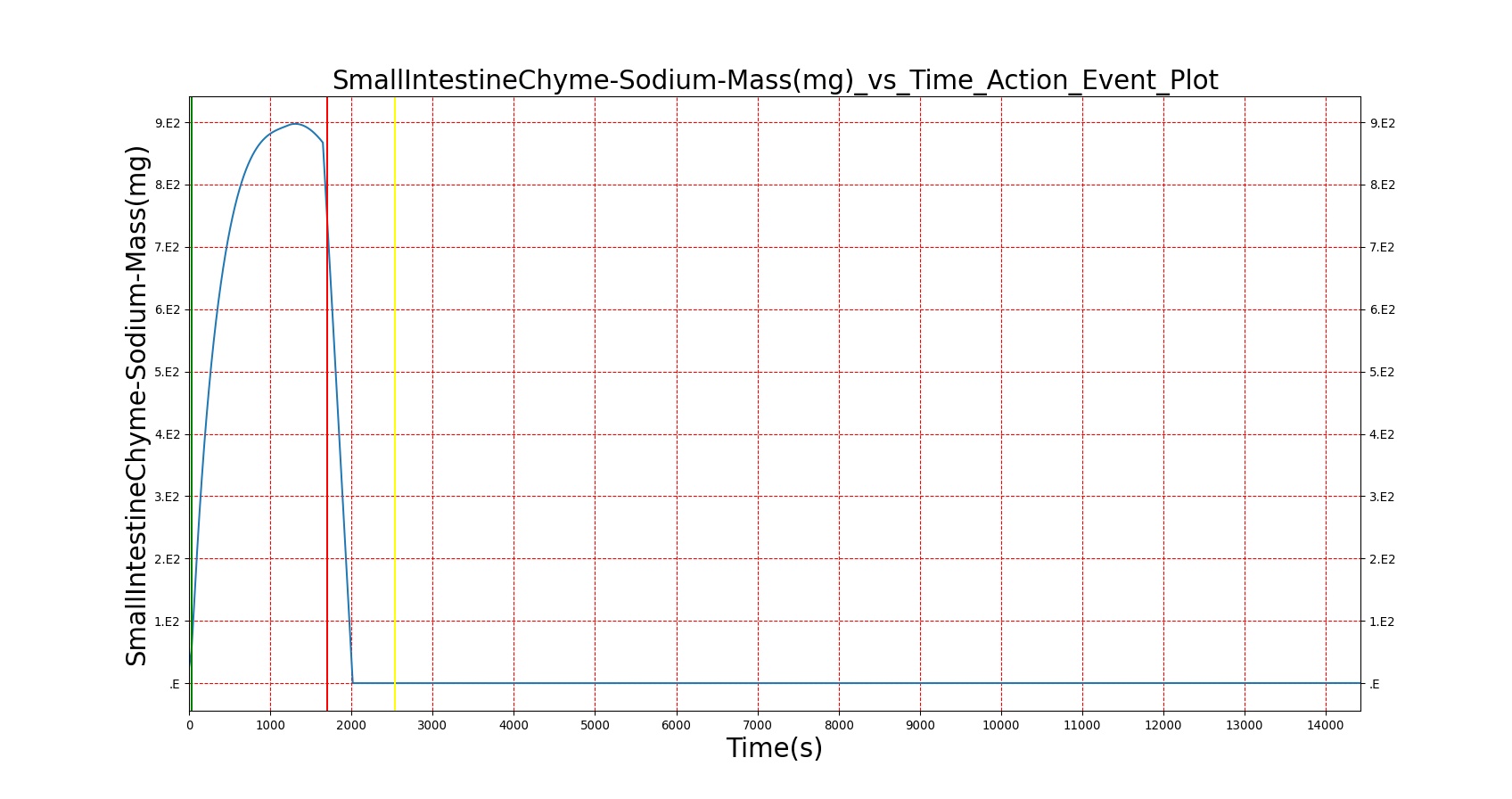
| 
|
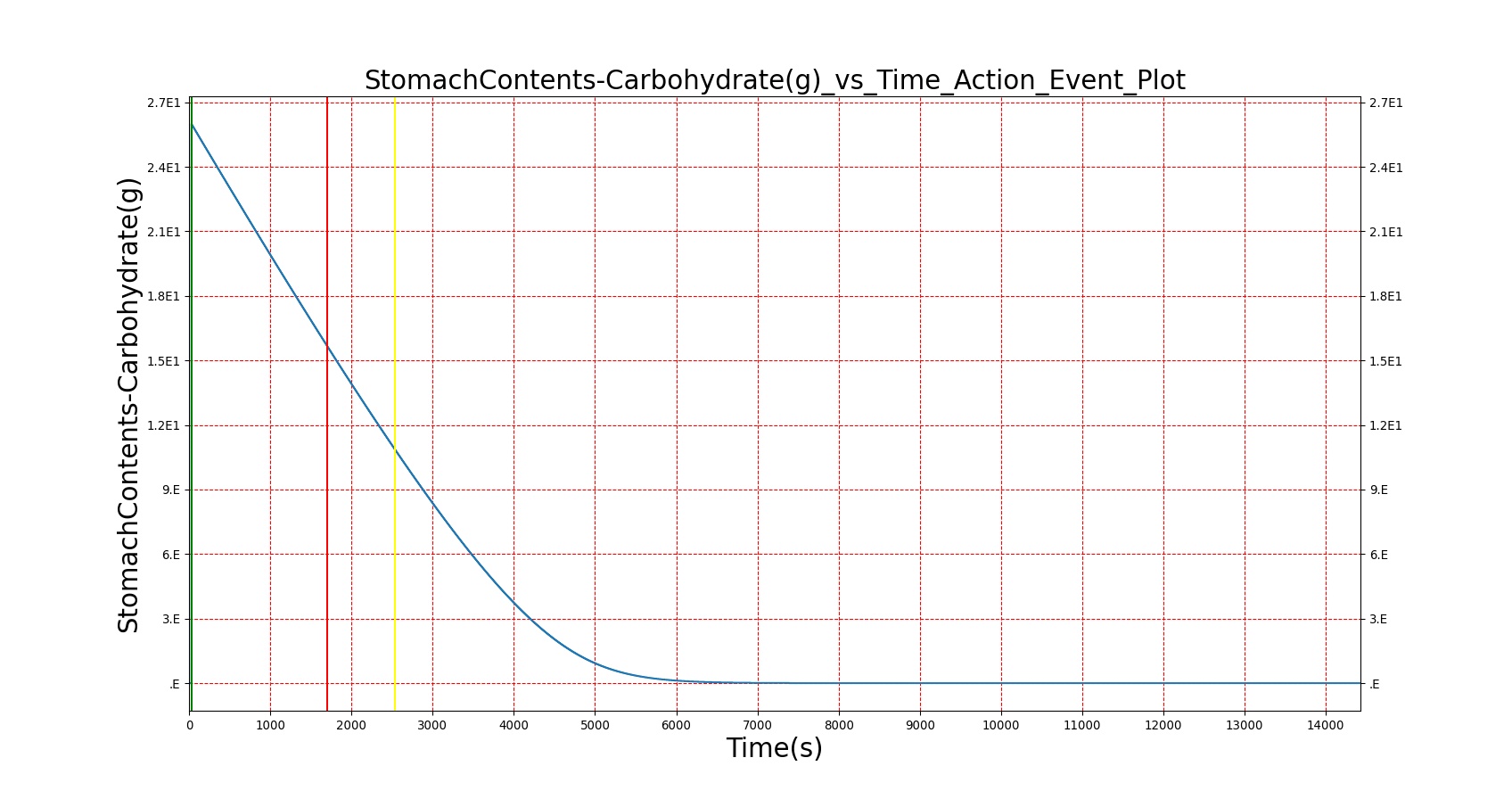
| 
|
Conclusions
The BioGears Gastrointestinal System lays a foundation for future efforts in modeling digestive physiology. The flexible definition of food allows for easy extensibility of the system by creating the ability for users to add to a library of meals.
Future Work
Coming Soon
The osmolarity of the digested material is not currently measured. Implementing this would allow for the creation of a more realistic osmolar gradient to determine the water absorption rate and gastric secretion. Permeability of the tissue matrix and substance concentration levels will be added to allow for actual diffusion of substances and fluid instead of active transport being implemented currently.
Recommended Improvements
- More Substances: By expanding the list of substances BioGears works with, the Gastrointestinal model will inherently gain fidelity as mechanisms are constructed to properly handle new inputs.
- Defecation: The Gastrointestinal system should eventually be able to handle undigestible material and the fluid loss due to defecation. This will also allow for a variety of disease states to be implemented.
- Vomiting: This can be added either as a symptom or an intervention to aid someone who has ingested a harmful substance.
Appendices
Acronyms
GI - Gastrointestinal
CV - Cardiovascular
Data Model Implementation
Cardiovascular
Gastrointestinal
Compartments
The GI system is integrated into the cardiovascular system, please refer to the compartement description associated with Cardiovascular Methodology.





